Page 16 - 《广西植物》2022年第10期
P. 16
1 6 3 4 广 西 植 物 42 卷
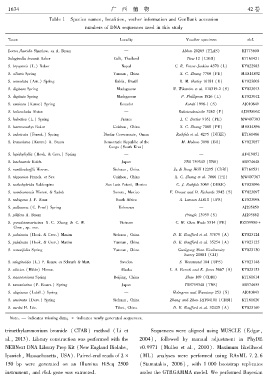
Table 1 Species namesꎬ localitiesꎬ vocher information and GenBank accession
numbers of DNA sequences used in this study
Taxon Locality Voucher specimen rbcL
Isoetes flaccida Shuttlew. ex A. Braun — Abbott 20265 (FLAS) KJ773600
Selaginella braunii Baker Cultꎬ Thailand Tiew 12 (CDBI) KT161421
S. bryopteris (L.) Baker Nepal C. R. Fraser ̄Jenkins 4370 (L) KY022983
S. ciliaris Spring Yunnanꎬ China X. C. Zhang 7780 (PE) MH814892
S. convoluta (Arn.) Spring Bahiaꎬ Brazil R. M. Harley 16181 (U) KY023003
S. digitata Spring Madagascar N. Wikström et al. 110319 ̄2 (S) KY023013
S. digitata Spring Madagascar P. Phillipson 1826 (L) KY023012
S. exaltata (Kunze) Spring Ecuador Korall 1996 ̄1 (S) AJ010849
S. helicoclada Alston — Rakotondrainibe 3262 (P) AJ295896C
S. helvetica (L.) Spring France J. C. Bertier 9161 (PE) MW407303
S. heterostachys Baker Guizhouꎬ China X. C. Zhang 7088 (PE) MH814896
S. imbricata (Forssk.) Spring Dhofar Governorateꎬ Oman Rothfels et al. 4275 (DUKE) KT161486
S. kraussiana (Kunze) A. Braun Democratic Republic of the M. Mokoso 3098 (BR) KY023057
Congo (South Kivu)
S. lepidophylla (Hook. & Grev.) Spring — — AF419051
S. lutchuensis Koidz. Japan TNS 759343 (TNS) AB574648
S. moellendorffii Hieron. Sichuanꎬ China Ju & Deng HGX 12295 (CDBI) KT161531
S. nipponica Franch. et Sav. Guizhouꎬ China X. C. Zhang et al. 7066 (PE) MW407367
S. nothohybrida Valdespino San Luis Potosíꎬ Mexico C. J. Rothfels 3069 (DUKE) KY023096
S. novoleonensis Hieron. & Sadeb Sonoraꎬ Mexico F. Drouet and D. Richards 3942 (S) KY023097
S. nubigena J. P. Roux South Africa A. Larsson AL810 (UPS) KY023098
S. pallescens (C. Presl) Spring Unknown — AJ295859
S. pilifera A. Braun — Pringle 13959 (S) AJ295862
S. pseudotamariscina X. C. Zhang & C. W. Vietnam C. W. Chen Wade 5314 (PE) MZ159980∗
Chenꎬ sp. nov.
S. pulvinata (Hook. & Grev.) Maxim Sichuanꎬ China D. E. Boufford et al. 37879 (A) KY023124
S. pulvinata (Hook. & Grev.) Maxim Yunnanꎬ China D. E. Boufford et al. 35254 (A) KY023125
S. remotifolia Spring Yunnanꎬ China Gaoligong Shan Biodiversity KY023130
Survey 21081 (GH)
S. selaginoides (L.) P. Beauv. ex Schrank & Mart. Sweden S. Weststrand 104 (UPS) KY023148
S. sibirica (Milde) Hieron. Alaska L. A. Viereck and K. Jones 5667 (S) KY023153
S. stauntoniana Spring Beijingꎬ China Zhao 169 (CDBI) KT161614
S. tamariscina (P. Beauv.) Spring Japan TNS759348 (TNS) AB574655
S. uliginosa (Labill.) Spring — Holmgren and Wanntorp 253 (S) AJ010843
S. uncinata (Desv.) Spring Sichuanꎬ China Zhang and Zhou DJY04101 (CDBI) KT161626
S. vardei H. Lév. Tibetꎬ China D. E. Boufford et al. 32425 (A) KY023169
Note: — indicates missing dataꎻ ∗ indicates newly generated sequences.
trimethylammonium bromide ( CTAB) method ( Li et Sequences were aligned using MUSCLE (Edgarꎬ
al.ꎬ 2013). Library construction was performed with the 2004)ꎬ followed by manual adjustment in PhyDE
NEBNext DNA Library Prep Kit (New England Biolabsꎬ v0.9971 ( Muller et al.ꎬ 2010). Maximum likelihood
Ipswichꎬ Massachusettsꎬ USA). Paired ̄end reads of 2 × (ML) analyses were performed using RAxML 7. 2. 6
150 bp were generated on an Illumina HiSeq 2500 (Stamatakisꎬ 2006)ꎬ with 1 000 bootstrap replicates
instrumentꎬ and rbcL gene was extracted. under the GTRGAMMA model. We performed Bayesian