Page 16 - 广西植物2024年1期
P. 16
1 2 广 西 植 物 44 卷
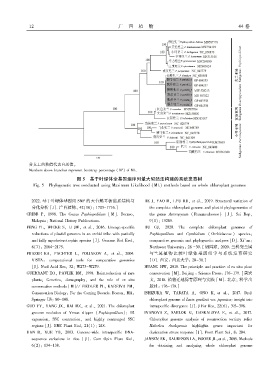
分支上的数值代表自展值ꎮ
Numbers above branches represent bootstrap percentage (BP) of ML.
图 5 基于叶绿体全基因组序列最大似然法构建的系统发育树
Fig. 5 Phylogenetic tree conducted using Maximum Likelihood (ML) methods based on whole chloroplast genomes
2022. 基于叶绿体基因组 SNP 的天台鹅耳枥谱系结构与 HE Jꎬ YAO Mꎬ LYU RDꎬ et al.ꎬ 2019. Structural variation of
分化分析 [J]. 广西植物ꎬ 42(10): 1703-1716.] the complete chloroplast genome and plastid phylogenomics of
CRIBB Pꎬ 1998. The Genus Paphiopedilum [ M]. Borneoꎬ the genus Asteropyrum ( Ranunculaceae) [ J]. Sci Repꎬ
Malaysia: National History Publications. 9(1): 15285.
FENG YLꎬ WICKE Sꎬ LI JWꎬ et al.ꎬ 2016. Lineage ̄specific HU GJꎬ 2020. The complete chloroplast genomes of
reductions of plastid genomes in an orchid tribe with partially Paphiopedilum and Cymbidium ( Orchidaceae ) species:
and fully mycoheterotrophic species [J]. Genome Biol Evolꎬ comparative genomic and phylogenetic analyses [D]. Xi’an:
8(7): 2164-2175. Northwest University: 28 -30. [胡国家ꎬ 2020. 兰科兜兰属
FRAZER KAꎬ PACHTER Lꎬ POLIAKOV Aꎬ et al.ꎬ 2004. 与兰属 植 物 比 较 叶 绿 体 基 因 组 学 与 系 统 发 育 研 究
VISTA: computational tools for comparative genomics [D]. 西安: 西北大学: 28-30.]
[J]. Nucl Acid Resꎬ 32: W273-W279. HUANG HWꎬ 2018. The principle and practice of ex situ plant
GUERRANT EOꎬ PAVLIK BMꎬ 1998. Reintroduction of rare conservation [M]. Beijing : Science Press: 176-179. [黄宏
plants: Geneticsꎬ demographyꎬ and the role of ex situ 文ꎬ 2018. 植物迁地保育原理与实践 [M]. 北京: 科学出
conservation methods [M] / / FIEDLER PLꎬ KAREIVA PMꎬ 版社: 176-179.]
Conservation Biology: For the Coming Decade. Bostonꎬ MA: ISHIZUKA Wꎬ TABATA Aꎬ ONO Kꎬ et al.ꎬ 2017. Draft
Springer US: 80-108. chloroplast genome of Larix gmelinii var. japonica: insight into
GUO YYꎬ YANG JXꎬ BAI MZꎬ et al.ꎬ 2021. The chloroplast intraspecific divergence [J]. J For Resꎬ 22(6): 393-398.
genome evolution of Venus slipper ( Paphiopedilum): IR IVANOVA Zꎬ SABLOK Gꎬ DASKALOVA Eꎬ et al.ꎬ 2017.
expansionꎬ SSC contractionꎬ and highly rearranged SSC Chloroplast genome analysis of resurrection tertiary relict
regions [J]. BMC Plant Biolꎬ 21(1): 248. Haberlea rhodopensis highlights genes important for
HAN Bꎬ XUE YBꎬ 2003. Genome ̄wide intraspecific DNA ̄ desiccation stress response [J]. Front Plant Sciꎬ 8: 204.
sequence variations in rice [ J]. Curr Opin Plant Biolꎬ JANSEN RKꎬ RAUBESON LAꎬ BOORE JLꎬet al.ꎬ 2005. Methods
6(2): 134-138. for obtaining and analyzing whole chloroplast genome